How to participate
![]()
How to ask for access, share your work...
COVID-19 Pulmonary Database
![]()
How to ask for access, share your work...
![]()
Data formats, conversion plugin...
![]()
How to connect to the plateform, download items...
Access to the database is restricted. Only a small portion of patient CT-scans are available on demand. To request access, one should registrer on the data platform and wait the approval from the project team. To registrer click here, please fill out the name of the institute.
As a large part of the database is not available to the public, participants in this project must agree to develop their applications only on the public part of the database and send the model to the management team for deployment on the secure part of the database. Participants must also agree to use the database only within the framework of this project Our team also undertakes not to use the shared models for any application other than this one.
The COVID-CTPRED clinical trial database contains pseudonymized data (images, clinical and
biological metadata)(“DATABASE”). The authorization to access and exploit the results
extracted and/or calculated from the DATABASE is conditional to the implicit agreement with
the following points:
1) Authorized users are not allowed to redistribute in any form the data from the
COVID-CTPRED database.
2) Citation rules when publishing any results obtained from the latter:
Any inquiries should be directed to the sponsor representative (Pr P.Croisille, Chairman of Radiology and Nuclear Medicine, CHU Saint-Etienne mail)
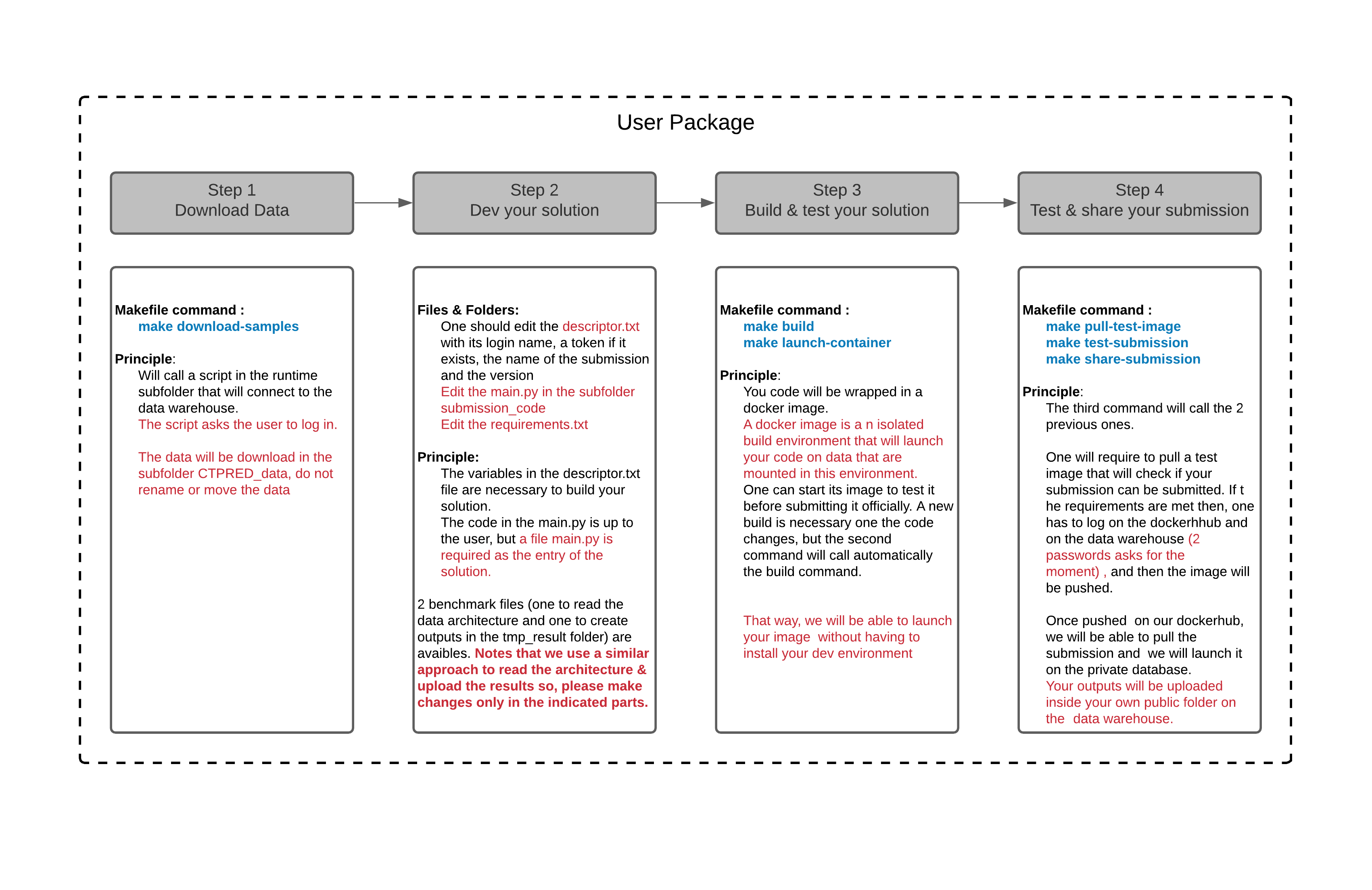
When you are registered in the warehouse platform, you have access to a package tool that will allow you to develop your solution.
This package provides a Makefile with different commands necessary to build and share your solution with us.
We are using Docker in order to create isolated environment for each solution.
You will have to download Docker locally in order to post what we called a image containing your solution.
We won't have access to your source code.
We implemented a Dockerhub that will host your submissions, you can, at any moment, ask us to remove your submissions.
The following graph gives the different steps required during the development of your solution.
Three formats are available
Upon request, a new set of parameters can be used to convert DICOM data to MHD format.
Once one's account is approved by the project team, one should log in to be able to see the database folder. To log in, must click on the 'Log In' button at the top right. If one does not have an account, create it with the button "Register", check How to participate FAQ.
Both viewers can be used independently from the data platform.
The first viewer based on the Itk-Vtk-Viewer library, also embedded in the data platform is simply used by uploading from your local environment the image you want to visualize.
The second viewer, Paraview-Glance, offers two different modes. You can use the viewer with local files or link the interface to a Girder Platform (such as the one we developp).
It appears to us that Nifti are well supported but other format as zip or MHD are not supported. Documentation can be found Here for itk-vtk-viewer or Here for Paraview-Glance .